October 21, 2007
My First PyMol Image
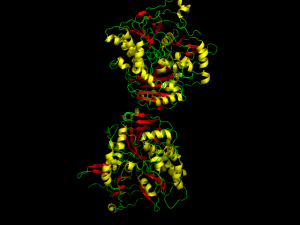
So eventually, if you want to do work with proteins, you have to actually look at some. I suppose it is inevitable. You will want to know the locations of alpha helices and beta sheets, you will want a sense of the way the thing all hangs together. These days, we don’t have to search through card catalogs of images of different proteins, painstakingly hand drawn by some poor soul to show the view that he thinks is most informative (but for your purposes blows goat chunks). Instead we can get files that detail the locations of each atom in the protein, and then generate the image automatically on our computer. Time to get a Molecular Visualizer.
I was faced with this task this weekend, as my Cell Biology and Biochemistry classes are quickly moving into the territory where I will be regularly required to look at the structures of proteins of interest. I came across 3 programs in my search (out of about 20 that I know of), all free. I’ll dally a bit with you here to discuss them.
Chimera was the first program that I downloaded. It appears to run fine on Mac OSX systems, though it is a X11 program, so devotees of the “Mac Look” may be disappointed when their cursor turns around and they are forced to interact with non-aqua rendering of windows. If this means nothing to you the take home message is: Its ugly.
Chimera probably works fine. I found it somewhat difficult to figure out immediately, as in, I did not look at any manuals, did not look up anything online, and frankly did not spend too much time on it. It appears to import pdb files from the Protein Database just fine, and may work better in this respect to the program I will discuss later, PyMol. I’m kinda a techy-linuxey kinda guy, so any high end program that doesn’t involve a text interface kind of frightens me. The though of spending arduous hours searching scroll down menu’s makes me shiver in my britches (to mix a metaphor). So Chimera now sits alone and abandoned in my applications folder, waiting for me to be neurotic enough to delete a 10 mb program in the search for extra space on my hard drive.
I also did some work with the KiNG viewer, a program made by the good people at Duke. KiNG obviously supports the “cartoon” view of proteins, but in the five minutes that I spent playing with it, I couldn’t get that feature to work. Furthermore, KiNG translates a pdb file into a so called kinemage (kinetic image), presumably to let you do cool and sophisticated things with it. But it also lacks a text interface and frankly, I didn’t like the feel of it too much. It is very likely that I would have gotten over this if I had honestly taken the time to learn to use it, which I may in the future. But I was already pretty taken with PyMol by this point.
On the upside, KiNG is a java application, which means that it is kind of half in and half out of the aqua interface. It is undoubtedly prettier than Chimera, so I guess if that is important to you, go for it.
As I have mentioned, the application that has won me over for the moment is PyMol. PyMol is made by Delano Scientific and it is the weird bastard son of people who put their faith early in open source but have become disenchanted.
According to their website, a high proportion of the images of proteins in journals like Nature and Science are made with PyMol. They pride themselves on the ability of their program to quickly make publication quality images.
Pymol comes in several flavors: Old, Expensive and Do-it-yourself. Pymol started as an open source project, but the developers have since begun charging for pre-compiled binaries the program for specific operating systems. You can get non-current versions of the software (0.99 is the latest freely available as of this writing) online for free, though you sacrifice in what ever new and improved features that they have added in the latest version, or whatever corrections to the drawing algorithms they may have made. If you want to pay money, you can become a subscriber, get the latest program, and support. If your school or institution is site licensed, you may be able to freely download it anyway. Will update this blog if PSU is so licensed. The do it yourself option is for those people who are willing to get their hands a little sweaty from some command line fu and linux trickery (not that sweaty).
Delano Scientific offers the source code for the latest versions of pymol for free. This means that you can download the source code, compile it on your own machine, and have a version of pymol specifically made for your own machine. (In the linux environment this individualization is preferred, but for the rest of us, it means that you cant just drag and drop the program onto your friend’s computer. ) If you want to do this, the easiest way is to install fink, a package manager program that will install linux apps onto your computer in a fairly automated way. If you install fink, simply enter:
fink install pymol-py25
on the command line, and presto! Your computer will make a lot of buzzing noises, the fan will spin, text will flash across your terminal interface and in about an hour, PyMol will be hidden away in the linux side of your shiny Macbox. If you want to do this, you need Xcode and X11, but those are free too.
You could simply go for PyMol 0.99, which runs in aqua and still has the text interface, but compared with the beauty of automatically compiling binaries on your own machine.
Anyway, I found pymol to be very straightforward. It has a somewhat non-intuitive UI, but once you get it (about 5 minutes for me) it became straightforward. It has a command line interface that I find very useful. I can enter:
load 2gmh.pdb
hide sticks
hide nonbonded
color yellow, ss h
color green, ss ""
color red, ss s
show cartoon
ray
png foo.png
and get the image at the start of this overly long entry. Furthermore, I was easily able to find good documentation for the program online, in the form of outdated user manuals for versions 0.99 and a community wiki. I have not been able to load the form of ATP synthase yet–there is some problem that I don’t understand. But other than that, I have been very pleased with the program. Four out of five stars.
UPDATED: I was using pdb file 1qo1 to try and play around with ATP synthase. This appears to simply be a file of atom locations, and not actually a file that will give you secondary structure. Thus when I loaded 1c17 and 1e79 (two other pdb files with full secondary structure data, I was able to move them both onto the 1oq1 file with the “align” command, and I was aces. PyMol all the way.
Updated 2: I have received information that PSU does not have a site license to pymol. Well shucks, guess you will have to stick with v 0.99 if you cant use Fink. If you can use fink, just type ‘fink install pymol-py25’ and you are golden. In the half hour that it takes to download and compile. So go get coffee.
Bertalan Meskó said,
October 21, 2007 at 7:23 pm
I’m looking forward to reading your posts! Thank you for the kind words about my blog and let me know if you need any kind of help.
I’m going to feature your blog in one of my next posts.
Berci
http://Scienceroll.com